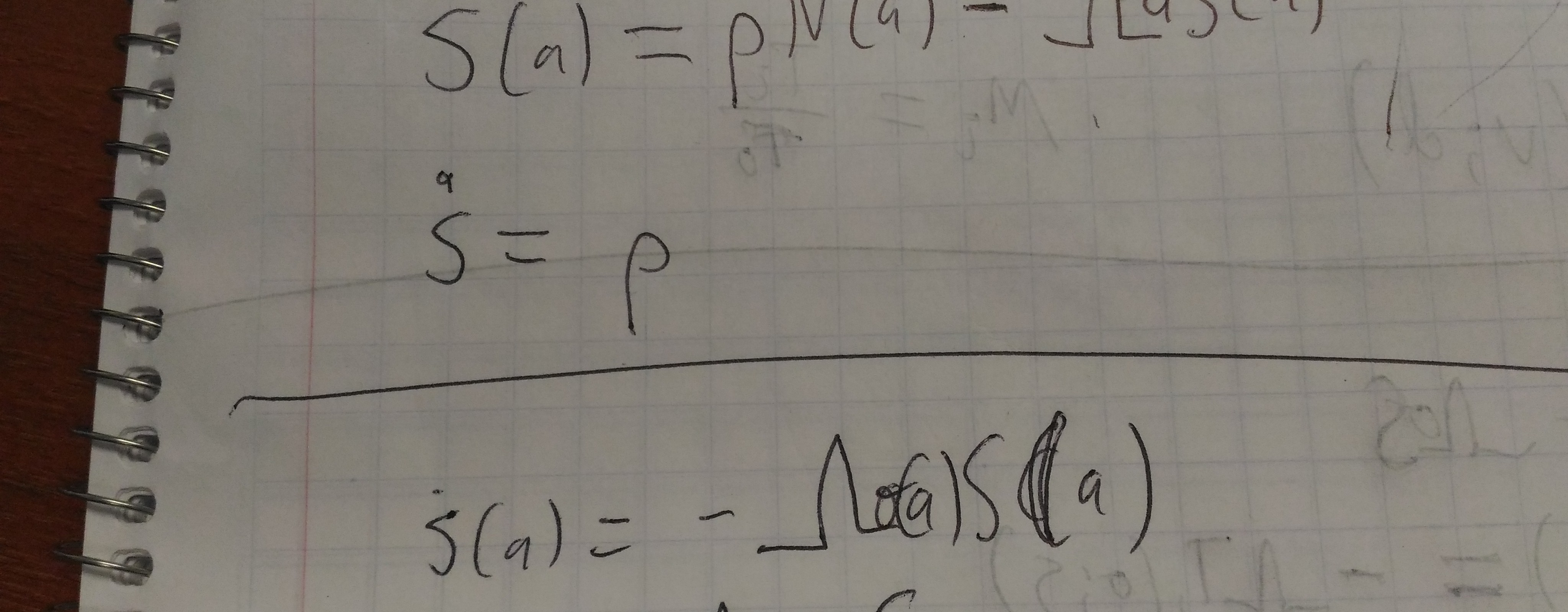Epidemic tSIR
Cara Brook introduced a very nice practical SIR experiment using cards to MMED. But the resulting data were very hard to analyze.
Summary
We assumed that the number of infectious and susceptibles were perfectly known; it therefore seems like it should be pretty easy to infer the one missing parameter (which corresponds to \(R_0\).
I encountered three problems:
- The log-link binomial approach which seems most natural is unstable. This is because the log link does not constrain the probability to be <1.
- This can be solved by using a
cloglogapproach; but see below.
- This can be solved by using a
lme4has a lot of trouble solving thecloglogmodel.- This seems to be due to initialization bugs and optimizer problems.
- In response to this problem, Ben Bolker has updated the development version of
lme4and provided some code with optimizer flags that allowslme4to fit these data
- The cards simulations don’t match the
cloglogassumptions. In particular, if we expect 26 infections in a population of 26, all of the susceptibles will be infected with probability 1, which would correspond to an infinitely high \(R_0\).- This matches the assumptions of the (unstable) log-link approach, but not those of the stable
cloglogapproach. - I don’t guess there is an easy way around it without spinning our own maximum likelihood. I did try adding some second-order terms to the cloglog; this helps a little.
- I have code which makes data that matches the hazard-based assumptions; the
cloglogmodel fits these perfectly, but can’t fit the data simulated with the real “cards” assumptions.
- This matches the assumptions of the (unstable) log-link approach, but not those of the stable